 This project is aimed at identifying the biological factors that drive the response of lotic microbial communities to massive organic and chemical pollutions, and at developing predictive tools of their ability to tolerate, resist, exploit pollutants and/or go back to original status (resilience) in river systems under anthropogenic pressure.
This project is aimed at identifying the biological factors that drive the response of lotic microbial communities to massive organic and chemical pollutions, and at developing predictive tools of their ability to tolerate, resist, exploit pollutants and/or go back to original status (resilience) in river systems under anthropogenic pressure.
For that purpose, a series of hypotheses on biodiversity-stability and biodiversity-functionality relationships in microbial communities will be initially formulated. They will be tested on model synthetic  communities of freshwater bacteria to which different stressors (input of organic matter, “invader” wastewater bacteria, and/or pollutants) will be applied in controlled conditions. Thereafter, the dynamics of the same communities will be analyzed in more complex systems (sterile, natural ecosystems) and eventually compared to in situ data. Theories and predictive tools that will emerge from this study will contribute to a better management of microbial communities and their processes in order to protect value and restore river ecosystem services.
communities of freshwater bacteria to which different stressors (input of organic matter, “invader” wastewater bacteria, and/or pollutants) will be applied in controlled conditions. Thereafter, the dynamics of the same communities will be analyzed in more complex systems (sterile, natural ecosystems) and eventually compared to in situ data. Theories and predictive tools that will emerge from this study will contribute to a better management of microbial communities and their processes in order to protect value and restore river ecosystem services.
DYNAMO partners: ESA-ULB (George, I, Servais, P. Goetghebuer, L), Applied Microbiology-UCL (Agathos, S)
Period of the study: July 2014- June 2022.
Financial support: FNRS – Fonds de la Recherche Scientifique (Belgium) (http://www.fnrs.be/)
ESA members participating to DYNAMO: George, I., Servais, P., Goetghebuer, L, Anzil, A., Bonal, M.
Publication:
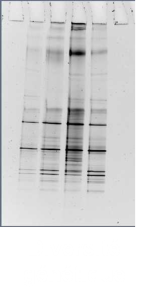
Goetghebuer, L., Bonal, M., Faust, K., Servais, P., George, I. 2019. The dynamic of a river model bacterial community in two different media reveals a divergent succession and an enhanced growth of most strains compared to monocultures. Microbial Ecology. 78 (2): 313-323
Goetghebuer, L., Servais, P. & George, I. 2017. Carbon utilization profiles of river bacterial strains facing sole carbon sources suggest metabolic interactions. FEMS Microbiology Letters. 364(10), fnx098. doi.org/10.1093/femsle/fnx098.
