Search for novel antibiotics in sponge-associated bacteria
In this project, we screen the microbiome of marine sponges for the discovery of compounds active against clinical bacterial strains. More specifically, we are focusing on the microbiome of Hymeniacidon perlevis and Halichondria panicea and searching for sponge bacteria inhibiting the growth of multi-resistant clinical strains. Two approaches are used in parallel: 1) a culture-dependent one: the screening of hundreds of strains that were isolated from the sponge tissue using classical or “revisited” cultivation media and 2) a culture-independent one: the cloning of large fragments of the microbiome’s DNA into various bacterial hosts and the subsequent screening of the metagenomic library.

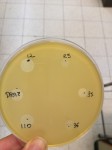
Figure 1: A piece of H. perlevis tissue (Picture by A. Rodriguez)
Figure 2: Results of an antagonism test.In the overlay, an inhibition zone is visible where the growth of S. aureus is prevented by the production of antibiotic molecule(s) by the underlying bacterial colony.(Picture by A. Rodriguez)
Period of the study: October 2017- September 2021
Partial financial support: CDR FNRS – Fonds de la Recherche Scientifique (Belgium) (http://www.fnrs.be/), project DISARM (“DIScovery of new Antibiotic molecules in the sponge micRobiome by heterologous expression in Multiple hosts”)
ESA members participating to the project: Rodriguez A., George I., Dechamps E.
